
|
Green fluorescent protein TurboGFP
- Bright green fluorescence
- Fast maturation at a wide range of temperatures
- High pH-stability and photostability
- Proven suitability to generate stably transfected cell lines
- Destabilized variant is available
- Recommended for gene expression analysis and cell and organelle labeling
 |
TurboGFP is an improved variant of the green fluorescent protein CopGFP cloned from copepod Pontellina plumata (Arthropoda; Crustacea; Maxillopoda; Copepoda) [Shagin et al., 2004]. It possesses bright green fluorescence (excitation/ emission max = 482/ 502 nm) that is visible earlier than fluorescence of other green fluorescent proteins.
TurboGFP is mainly intended for applications where fast appearance of bright fluorescence is crucial. It is specially recommended for cell and organelle labeling and tracking the promoter activity. Destabilized TurboGFP variant allows accurate analysis of rapid and/or transient events in gene regulation.
|
Main properties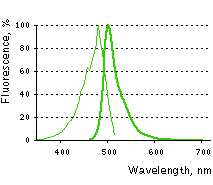
TurboGFP normalized excitation (thin line) and emission (thick line) spectra.
Spectra viewer tool
Download TurboGFP spectra (xls)
| | CHARACTERISTIC | |
|---|
* Brightness is a product of extinction coefficient and quantum yield, divided by 1000.
| | Molecular weight, kDa | 26 | | Polypeptide length, aa | 232 | | Fluorescence color | green | | Excitation maximum, nm | 482 | | Emission maximum, nm | 502 | | Quantum yield | 0.53 | | Extinction coefficient, M-1cm-1 | 70 000 | | Brightness* | 37.1 | | Brightness, % of EGFP | 112 | | pKa | 5.2 | | Structure | dimer | | Aggregation | no | | Maturation rate at 37°C | super fast | | Photostability | high | | Cell toxicity | not observed | | Main advantages | superbright and fast-maturing green fluorescent protein | | Possible limitations | dimer, limited applicability for fusions generation |
|
|---|
Recommended filter sets and antibodies
TurboGFP can be recognized using Anti-TurboGFP(d) (Cat.# AB513) antibody available from Evrogen.
TurboGFP can be detected using common fluorescence filter sets for EGFP, FITC, and other green dyes. Recommended Omega Optical filter sets are QMAX-Green, XF100-2, XF100-3, XF115-2, and XF116-2.
Performance and use
TurboGFP can be expressed and detected in a wide range of organisms including cold-blooded animals. Mammalian cells transiently transfected with TurboGFP expression vectors produce bright fluorescence in 8-10 hrs after transfection. No cytotoxic effects or visible protein aggregation are observed. TurboGFP can be used in multicolor labeling applications with blue, true-yellow, red, and far-red fluorescent dyes.
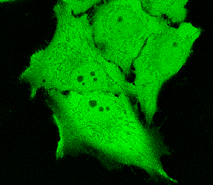 | TurboGFP expression in transiently transfected mammalian cells.
|
|---|
TurboGFP suitability to generate stably transfected cells has been proven by Marinpharm company.
Despite its dimeric structure, TurboGFP performs well in some fusions. However, for protein labeling applications we recommend using specially optimized monomeric TagFPs.
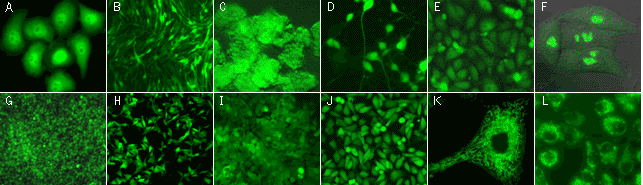 | |
|---|
TurboGFP expression in stably transfected mammalian cell lines.
(A) Human cervix carcinoma HeLa cells with TurboGFP in cytoplasm; (B) C2C12 mouse myoblasts with TurboGFP in cytoplasm; (C) PC-12 rat phaeochromocytoma cells with TurboGFP in cytoplasm; (D) PC-12 rat phaeochromocytoma cells with TurboGFP in cytoplasm after the addition of nerve growth factor (cells differentiate irreversibly into neuron-like cells); (G) Walker 256 rat tumour cells with TurboGFP in cytoplasm; (H) M3-mouse melanoma with TurboGFP in cytoplasm; (I) 3T3 mouse fibroblast with TurboGFP in cytoplasm; (J) T24 human bladder carcinoma with TurboGFP in cytoplasm; (E) Chinese Hamster Ovary Cells CHO-K1 with TurboGFP in cytoplasm; (F) HeLa cells expressing TurboGFP-fibrillarin fusion; (K) HeLa cells expressing mitochondria-targeted TurboGFP; (L) T24 human bladder carcinoma expressing mitochondria-targeted TurboGFP.
Images were kindly provided by Dr. Christian Petzelt (Marinpharm).
|
TurboGFP maturation kinetics: TurboGFP allows monitoring the activity from early promoters. It matures noticeably faster than EGFP and most other fluorescent proteins. This difference in performance is illustrated here using both in vitro analysis of TurboGFP and EGFP refolding and maturation kinetics and in vivo examination of the developing Xenopus embrios expressing either TurboGFP or EGFP.
Refolding and maturation kinetics of TurboGFP and some other fluorescent proteins in vitro| Fluorescent protein | Refolding
half-time, s | Maturation
half-time,s | kox
(10-4s-1) |
Reference |
|---|
| Samples of fluorescent proteins were heated to 95°C in denaturation solution (8 M urea, 1 mM DTT) for 4 min. Refolding reactions were initiated upon 100-fold dilution into the renaturation buffer (35 mM KCl, 2 mM MgCl2, 50 mM Tris pH 7.5, 1 mM DTT). In maturation assay, 5 mM freshly dissolved dithionite was added to the denaturation solution [Reid and Flynn, 1997]. Due to the instability of dithionite at high temperatures, to provide for complete chromophore reduction the sample was cooled to 25°C and the addition of 5 mM dithionite followed by heating to 5°C were repeated. Protein refolding and maturation were followed by measuring the recovery of fluorescence using Varian Cary Eclipse Fluorescence Spectrophotometer, chamber temperature maintained at 25°C. Maturation rate constants (kox) were determined by computer-fitting the kinetic data to the first order exponential decay (Origin 6.0). | | EGFP | 90.6 | 3915 | 1.77 | Evdokimov et al., 2006 | | Venus | 46.2 | 4076 | 1.70 | Kremers et al., 2006 | | SYFP2 | 69.3 | 3300 | 2.10 | Kremers et al., 2006 | | TurboGFP | 11.0 | 1468 | 4.72 | Evdokimov et al., 2006 | 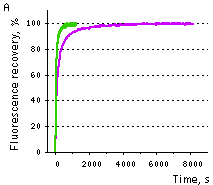 | 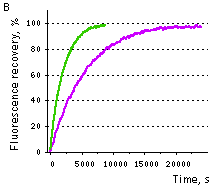 | Comparison of EGFP (violet lines) and TurboGFP (green lines) refolding and maturation speed in vitro.
Normalized fluorescence recovery plots are shown.
(A) — refolding kinetics;
(B) — chromophore maturation kinetics.
|
|---|
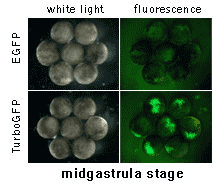 | 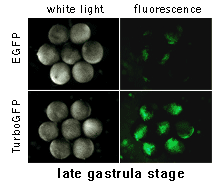 | In vivo comparison of TurboGFP and EGFP maturation in developing Xenopus embryos
Vectors encoding TurboGFP and EGFP under the control of CMV promoter were microinjected into animal poles of Xenopus embryos at the stage of two blastomeres. Living embryos were then photographed from the animal pole at the middle and late gastrula stages.
Experimental data were presented by Dr. A. Zaraisky, Institute of Bioorganic Chemistry, RAS (Moscow, Russia).
|
|---|
Available variants and fusions
| Variant | Description | Related vector | Cat.# | Click for image |
|---|
 |
|
Humanized TurboGFP
|
TurboGFP codon usage is optimized for high expression in mammalian cells [Haas et al., 1996], but it can be successfully expressed in many other heterological systems.
|
pTurboGFP-C
|
FP511
|
|
pTurboGFP-N
|
FP512
|
|
pTurboGFP-B
|
FP513
|
|
pTurboGFP-PRL
|
FP515
|
|
Gateway® TurboGFP-C entry clone
|
FP521
|
|
Gateway® TurboGFP-N entry clone
|
FP522
|
|
Destabilized TurboGFP variant (TurboGFP-dest1)
|
TurboGFP-dest1 is produced by fusing the initial protein with PEST amino acid sequence encoded by region 422-461 of mouse ornithine decarboxylase gene [Li et al., 1998]. This sequence targets the protein to degradation and enables a rapid protein turnover. TurboGFP-dest1 retains spectral properties of the initial protein, but has shorter half-life (approximately 2 hrs) as measured by the analysis of fluorescence intensity of cells treated with a protein synthesis inhibitor, cycloheximide. Because of rapid turnover, TurboGFP-dest1 can be used to measure changes in gene expression.
|
peTurboGFP-PRL-dest1
|
FP523
|
|
peTurboGFP-dest1
|
FP524
|
|
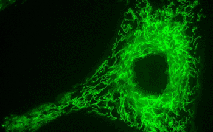
TurboGFP-mito fusion
|
A mitochondrial targeting sequence (MTS) is fused to the TurboGFP N-terminus. MTS was derived from the subunit VIII of human cytochrome C oxidase [Rizzuto et al., 1989; Rizzuto et al., 1995]. When expressed in mammalian cells, this variant provides green fluorescent labeling of mitochondria.
|
pTurboGFP-mito
|
FP517
|
|
References:
-
Evdokimov AG, Pokross ME, Egorov NS, Zaraisky AG, Yampolsky IV, Merzlyak EM, Shkoporov AN, Sander I, Lukyanov KA, Chudakov DM.
Structural basis for the fast maturation of Arthropoda green fluorescent protein.
EMBO Rep. 2006; 7 (10):1006-12. / pmid: 16936637
-
Haas J, Park EC, Seed B.
Codon usage limitation in the expression of HIV-1 envelope glycoprotein.
Curr Biol. 1996; 6 (3):315-24. / pmid: 8805248
-
Kremers GJ, Goedhart J, van Munster EB, Gadella TW.
Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius.
Biochemistry. 2006; 45 (21):6570-80. / pmid: 16716067
-
Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR.
Generation of destabilized green fluorescent protein as a transcription reporter.
J Biol Chem. 1998; 273 (52):34970-5. / pmid: 9857028
-
Reid BG, Flynn GC.
Chromophore formation in green fluorescent protein.
Biochemistry. 1997; 36 (22):6786-91. / pmid: 9184161
-
Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T.
Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells.
Curr Biol. 1995; 5 (6):635-42. / pmid: 7552174
-
Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, DiMauro S, Schon EA.
A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues.
J Biol Chem. 1989; 264 (18):10595-600. / pmid: 2543673
-
Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, Widder EA, Lukyanov SA, Matz MV.
GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity.
Mol Biol Evol. 2004; 21 (5):841-50. / pmid: 14963095
|













