
|
||||||||||
|
||||||||||

|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The vector sequence has been compiled using the information from sequence databases, published literature, and other sources, together with partial sequences obtained by Evrogen. This vector has not been completely sequenced. |
|||||||||||||||||||||||||||||||||||||||||||||||||
 Download
|
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Nhe I | Bgl II | Sac I | Hind III | EcoR I | Sal I | Kpn I | Apa I | BamH I | Age I | SLIM | |||||||||||||||||
| Afe I | Xho I | Pst I | Sac II* | ||||||||||||||||||||||||
| GCT | A.GC | G.CT | A.CCG.GAC.TC | A.GAT. | CT | C. | GAG. | CTC. | AAG.CTT. | C | GA.ATT. | C | TG.CA | G. | TCG.AC | G.GTA. | CC | G.C | GG. | G | CC.C | G | G.G | AT.CC | A.CCG.GT | C.GCC.ACC. | ATG.GTG.AGC |
* – not unique site.
Vector description
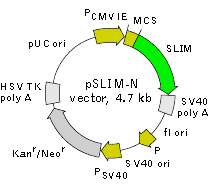
pSLIM-N is a mammalian expression vector encoding green fluorescent protein SLIM (see reporter description). The vector allows generation of fusions to the SLIM N-terminus and expression of SLIM fusions or SLIM alone in eukaryotic (mammalian) cells.
SLIM codon usage is optimized for high expression in mammalian cells (humanized) [Haas et al., 1996]. To increase mRNA translation efficiency, Kozak consensus translation initiation site is generated upstream of the SLIM coding sequence [Kozak, 1987]. Multiple cloning site (MCS) is located between PCMV IE and SLIM coding sequence.
The vector backbone contains immediate early promoter of cytomegalovirus (PCMV IE) for protein expression, SV40 origin for replication in mammalian cells expressing SV40 T-antigen, pUC origin of replication for propagation in
SV40 early promoter (PSV40) provides neomycin resistance gene (Neor) expression to select stably transfected eukaryotic cells using G418. Bacterial promoter (P) provides kanamycin resistance gene expression (Kanr) in
Generation of SLIM fusion proteins
A localization signal or a gene of interest can be cloned into MCS of the vector. It will be expressed as a fusion to the SLIM N-terminus when inserted in the same reading frame as SLIM and no in-frame stop codons are present. The inserted sequence should contain an initiating ATG codon. SLIM-tagged fusions retain fluorescent properties of the native protein allowing fusion localization in vivo. Unmodified vector will express SLIM when transfected into eukaryotic (mammalian) cells.
Note: The plasmid DNA was isolated from dam+-methylated
Expression in mammalian cells
pSLIM-N vector can be transfected into mammalian cells by any known transfection method. CMV promoter provides strong, constitutive expression of SLIM or its fusions in eukaryotic cells. If required, stable transformants can be selected using G418 [Gorman, 1985].
Propagation in
Suitable host strains for propagation in
Location of features
PCMV IE: 1-589
Enhancer region: 59-465
TATA box: 554-560
Transcription start point: 583
MCS: 592-678
BrUSLEE
Kozak consensus translation initiation site: 672-682
Start codon (ATG): 679-681
Stop codon: 1396-1398
SV40 early mRNA polyadenylation signal
Polyadenylation signals: 1551-1556 & 1580-1585
mRNA 3' ends: 1589 & 1601
f1 single-strand DNA origin: 1648-2103
Bacterial promoter for expression of Kanr gene
-35 region: 2165-2170
-10 region: 2188-2193
Transcription start point: 2200
SV40 origin of replication: 2444-2579
SV40 early promoter
Enhancer (72-bp tandem repeats): 2277-2348 & 2349-2420
21-bp repeats: 2424-2444, 2445-2465 & 2467-2487
Early promoter element: 2500-2506
Major transcription start points: 2496, 2534, 2540 & 2545
Kanamycin/neomycin resistance gene
Neomycin phosphotransferase coding sequences:
Start codon (ATG): 2628-2630
Stop codon: 3420-3422
G->A mutation to remove Pst I site: 2810
C->A (Arg to Ser) mutation to remove BssH II site: 3156
Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal
Polyadenylation signals: 3658-3663 & 3671-3676
pUC plasmid replication origin: 4007-4650
References:
- Gorman C. High efficiency gene transfer into mammalian cells. In DNA cloning: A Practical Approach, Vol. II. Ed. D. M. Glover. (IRL Press, Oxford, U.K.). 1985; 143-90.
- Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996; 6 (3):315-24. / pmid: 8805248
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987; 15 (20):8125-48. / pmid: 3313277
Notice to Purchaser:
SLIM-related materials (also referred to as "Products") are intended for research use only.
|
Copyright 2002-2023 Evrogen. All rights reserved. Evrogen JSC, 16/10 Miklukho-Maklaya str., Moscow, Russia, Tel +7(495)988-4084, Fax +7(495)988-4085, e-mail:evrogen@evrogen.com |


